Extended-spectrum beta lactamase and carbapenemaseproducing Klebsialla spp. in urine and fecal samples obtained from hospitals and communities in Lagos, Nigeria
Keywords:
Extended Spectrum Beta-Lactamase, Urinary Tract Infections, Carbapenemase, Klebsiella sppAbstract
The use of beta-lactams has tremendously increased since the discovery of antibiotics. This has led to the emergence of certain resistant genes such as Extended Spectrum Beta-Lactamase (ESBL) which confer resistance to third generation cephalosporins. The objective of this study was to determine the prevalence of Extended Spectrum Beta Lactamase (ESBL) genes, Carbapenem resistance genes (blaKPC, blaOXA and IMP) and outer membrane porins genes (OMP-35, OMP-36, and OMP-36N) from different hospitals and laboratories in Ikeja-Lagos, Nigeria. A total of 177 bacterial isolates were collected between May 2017 to July 2017 from patients with urinary tract infections (UTI) and gastroenteritis. They were identified biochemically and investigated for ESBL and Carbapenemase
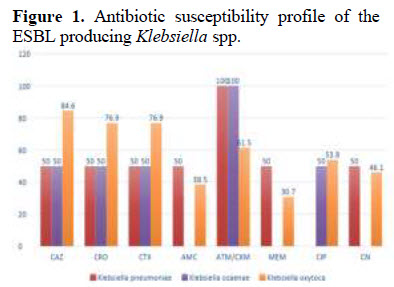
production using phenotypic Double Disk Synergy Test (DDST) and Modified Hodges’ Test respectively. Antibiotics susceptibility profile was also investigated. Multiplex PCR was used to detect the genes responsible for the resistant genes. Out of 177 bacterial isolates, 47 (26.6%) were identified as Klebsiella spp and 17 (36.1%) were ESBL positive and then 5 (29.4%) were positive for carbapenem resistance. Multiplex PCR revealed that 3 (27.3%) possessed both blaCTX-M and blaSHV genes, 6 (54.5%) possessed only blaCTX-M gene while only 2 (18.2%) possessed only blaSHV gene. Also, 13 (76.5%) possessed only blaKPC gene. However, blaTEM as well as IMP, OXA-48, OMP 35, OMP 36 and OMP 36N genes were not detected. This study revealed that antibiotic resistance is on the rise and preventive measures should be put in place by both government and health care providers to curtail this trend.
References
Abdullah, E., Syeda, S., Waseema, A. & Fatima, S. (2015). Identification of Klebsiella pneumoniae and Klebsiella oxytoca in urine specimens from a laboratory in Karachi. Microbiology Research International, 3(3), 37 - 40.
Ambler, R. P. (1980). The structure of b-lactamases. Philosophical Transactions of the Royal Society of London B Biological Sciences, 289, 321–331.
Anne, M. Q and Karen, B. (2007). Carbapenemases: the versatile beta-lactamases. Clinical Microbiology Review, 20(3), 440-458.
Bharat, P. M., Janak, K., Rajan, D. K., Shyam, M. K., Prem, K. K., & Tuladhar, N. R. (2006). Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. International Journal of Infectious Diseases, 10, 434-438.
Bonnet, R. (2004). Growing group of Extended-Spectrum β-lactamases: the CTX-M enzymes. Antimicrobial Agents and Chemotherapy, 48,
-14.
Bora, A., Hazarika, N. K., Shukla, S. K., Prasad, K. N., Sharma, J. B., Ahmed, G. (2014). Prevalence of blaTEM, blaSHV and blaCTX-M genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India. Journal of Pathology and Microbiology, 57(2), 249-254.
Bradford, P. (2001). Extended-spectrum betalactamases in the 21st century: characterization, epidemiology, and detection of this important
resistance threat. Clinical Microbiology Revolution, 14(4): 933-51.
Bush, K. and Jacoby, G. (2010). An updated functional classification of b-lactamases. Antimicrobial Agents and Chemotherapy, 54(3), 969–976.
Bush, K., Jacoby, G. A., Medeiros, A. A. (1995). A functional classification scheme for betalactamases and its correlation with molecular structure. Antimicrobial Agents and Chemotherapy, 39 (6): 1211–33.
Clinical and Laboratory Standards Institute. (2012). Performance standards for antimicrobial susceptibility testing; 22nd informational
supplement. CLSI document M100-S22 Clinical and Laboratory Standards Institute, Wayne, PA.
Datta, N., & Kontomichalou, P. (1965). Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature International Journal, 208, 239–241.
Garrec, H., Drieux-Rouzet, L., Golmard, J., Jarlier, V. & Robert, J. (2011). Comparison of nine phenotypic methods for detection of extendedspectrum beta-lactamase production by Enterobacteriaceae. Journal of Clinical Microbiology, 49, 1048 - 1057.
Gazouli, M., Tzelepi, E., Markogiannakis, A., Legakis, N. J., & Tzouvelekis, L. S. (1998). Two novel plasmid-mediated cefotaxime hydrolyzing
b-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. Oxford University Press, University of Oxford. FEMS Microbiology Letters, 165, 289–293.
Livermore, D. M. (1995). Beta-Lactamases in laboratory and clinical resistance. Clinical Microbiology Revolution, 8(4), p. 557-84.
Maninder, K., & Aruna, A. (2013). Occurrence of the CTX-M, SHV and the TEM genes among the Extended Spectrum Beta- Lactamase producing isolates of Enterobacteriaceae in a Tertiary Care in Hospital of North India. Journal of Clinical and Diagnostic Research, 7(4),
-646.
Masoume, B., Abazar, P., Shiva, M., Malihe, T. & Gholamreza, I. (2015). Detection of the Klebsiella pneumoniae Carbapenemase (KPC) in K. pneumoniae Isolated from the Clinical Samples by the Phenotypic and Genotypic Methods. Iranian Journal of Pathology, 10 (3): 199-205.
Paterson, D. L., & Bonomo, R. A. (2005). Extended-spectrum β-lactamases: a clinical update. Clinical Microbiology Reviews, 18(4), 657-86.
Paterson, D. L. & Bonomo, R. A. (2005). Extendedspectrum beta lactamases: a clinical update. Clinical Microbiology Revolution, 18, 657-686.
Pitout, J., Hossain, A. & Hanson, N. (2004). Phenotypic and molecular detection of CTX-Mbeta-lactamases produced by Escherichia coli and Klebsiella spp. Journal of Clinical Microbiology, 42, 5715 - 5721.
Rabindra, N. & Sibanarayan, R. (2014). Monitoring invitro Antibacterial Efficacy of twenty six Indian Spices against Multidrug Resistant
Urinary Tract Infecting Bacteria. Integrative Medicine Research, 3(3), 133-141.
J. J. (2000). Extended Spectrum Beta-Lactamase: How big is the problem? Clinical Microbiology and Infection, 6(2), 2-6.
Shakil, S., Ali, S., Akram, S. & Khan, A. (2010). Risk Factors for Extended-Spectrum Beta-Lactamase Producing Escherichia coli and Klebsiella pneumoniae Acquisition in a Neonatal Intensive Care Unit. Journal of Tropical Pediatrics, 56 pg 2.
Shakti, R., Debasmita, D., Mahesh, C., Sahu, R. & Padhy, N. (2014). Surveillance of ESBL producing Multidrug Resistant Escherichia coli in a teaching hospital in India. Asian Pacific Journal of Tropical Diseases, 4 (2), 140 - 149.
Soughakoff, W., Goussard, S., & Courvalin, P. (1998). TEM-3 beta lactamases which hydrolyzes broad-spectrum cephalosporins is derived from the TEM-2 penicillinases by two amino acid substitutions. FEMS Microbiology Letters, 56, 343–348.
Tzouvelekis, L.S., & Bonomo, R.A. (1999). SHVtype b-lactamases. Current Pharmacological Descriptions, 5, 847–864.
Tzouvelekis, L.S., Tzelepi, E., Tassios, P.T., & Legakis, N.J., (2000) CTX-M type sbetalactamases: an emerging group of Extended Spectrum enzymes. International Journal of Antimicrobial Agents, 14, 137–143.
Walsh, C. (2003). Antibiotics: actions, origins, resistance. The American Society for Microbiology (ASM Press). Washington, DC.