Biotechnological Potential of Marine-Derived Fungi for Textile Dye Degradation via Laccase-Like Activity
DOI:
https://doi.org/10.53848/ssstj.v12i2.974Keywords:
Meyerozyma guilliermondii, Penicillium oxalicum, Marine fungi, Laccase-like activity, Dyes, OxidativeAbstract
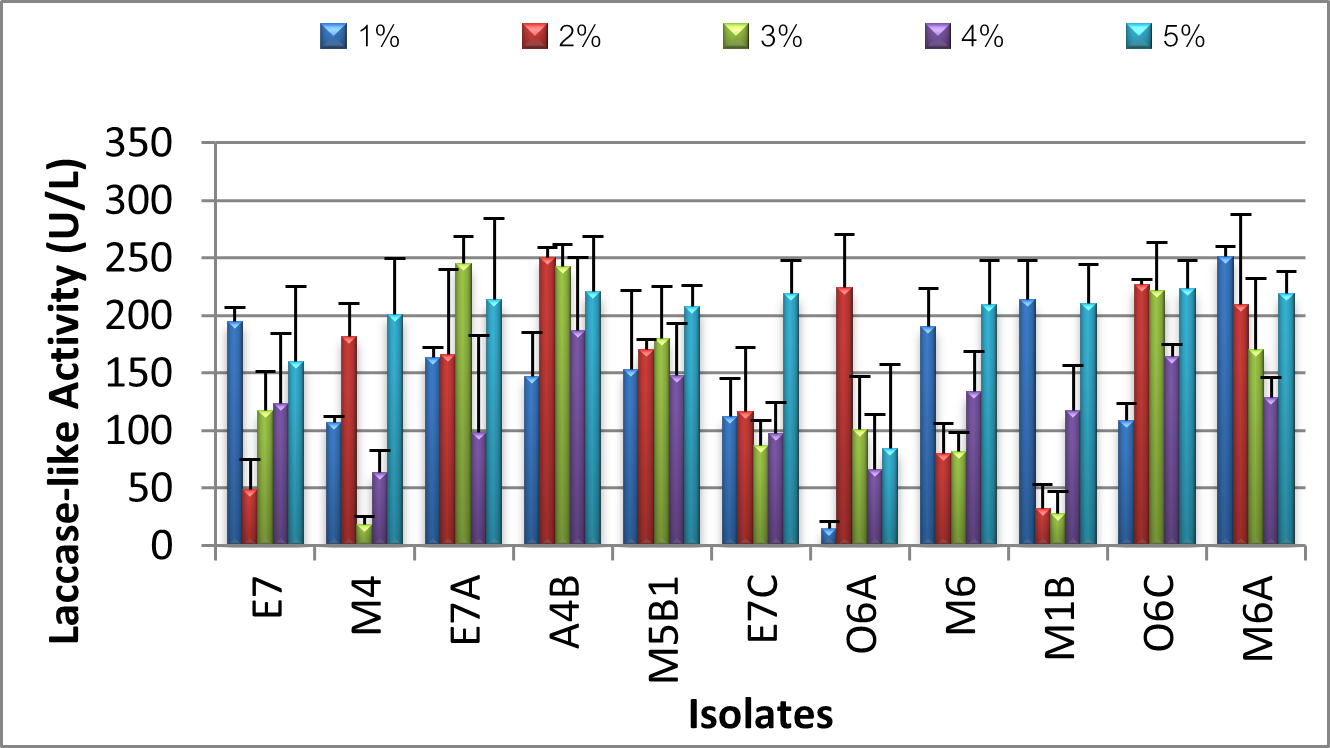
The complex structure of synthetic dyes has made them challenging to decolorize, despite their widespread use in a variety of industries, including textiles, cosmetics, printing, paper, and pharmaceuticals. Enzymes such as oxidoreductases and peroxidases are employed in novel biotechnological procedures. An affordable and environmentally friendly solution to the issue of decolorizing commercial dyes is the use of oxidative enzymes, such as laccases derived from fungi. This study screened marine-derived fungal strains isolated from five coastal areas in Lagos, Nigeria, in order to identify laccase-like activities and assess the isolates' ability to decolorize industrial dyes in solid and liquid media by monitoring the radial growth, percentage decolorization, biosorption, and laccase activity after seven days of incubation with the dyes. After several steps of culturing on potato dextrose agar (PDA) plates, forty pure fungal cultures were isolated from a total of 100 samples (wood, nets, plants, clothing materials, soil, and water) collected from five different marine biotopes in Lagos: Oniru Beach, Makoko Lagoon, Elegushi Beach, Adekunle Lagoon, and Unilag waterfront. Through preliminary oxidative ability screening with 2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), eleven isolates were discovered. Six strains of Meyerozyma guilliermondii, two strains of Rhodotorula mucilaginosa, one strain of Candida tropicalis, and two strains of Penicillium oxalicum were identified as belonging to the following species by means of molecular-based taxonomic approaches. Meyerozyma guilliermondi A4B and Penicillium oxalicum M6A effectively decolorized four synthetic dyes from various dye families and demonstrated the highest level of oxidative capacity and grew best at 2 mM CuSO4 as an inducer and 2% and 1% NaCl to mimic a marine environment, and both produced the highest laccase-like active cell-free supernatants activity.
References
Ali, B., Al-Wabel, N. A., Shams, S., Ahamad, A., Khan, S. A., & Anwar, F. (2015). Essential oils used in aromatherapy: A systemic review. Asian Pacific Journal of Tropical Biomedicine, 5(8), 601–611. https://doi.org/10.1016/j.apjtb.2015.05.007
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402. https://doi.org/10.1093/nar/25.17.3389
Amend, A., Burgaud, G., Cunliffe, M., Edgcomb, V. P., Ettinger, C.L., Gutiérrez, M. H., Heitman, J., Hom, E. F. Y., Ianiri, G., Jones, A. C., Kagami, M., Picard, K. T., Quandt, C. A., Raghukumar, S., Riquelme, M., Stajich, J., Vargas-Muñiz, J., Walker, A. K., Yarden, O., & Gladfelter, A. S. (2019). Fungi in the marine environment: Open questions and unsolved problems. mBio, 10, e01189-18.https://doi.org/10.1128/mbio.01189-18
Atalla, M. M., Zeinab, H. K., Eman, R. H., Amani, A. Y., & Abeer, A. A. (2013). Characterization and kinetic properties of the purified Trematosphaeria mangrovei laccase enzyme. Saudi Journal of Biological Sciences, 20(4), 373–381. https://doi.org/10.1016/j.sjbs.2013.04.001
Bankole, P. O., Omoni, V. T., Mulla, S. I., Adebajo, S. O., & Adekunle, A. A. (2022). Co-biomass degradation of fluoranthene by marine-derived fungi; Aspergillus aculeatus and Mucor irregularis: Comprehensive process optimization, enzyme induction and metabolic analyses. Arabian Journal of Chemistry, 15, 104036. https://doi.org/10.1016/j.arabjc.2022.104036
Ben Ali, W., Chaduli, D., Navarro, D., Lechat, C., Turbé-Doan, A., Bertrand, E., Faulds, C. B., Sciara, G., Lesage-Meessen, L., Record, E., & Mechichi, T. (2020). Screening of five marine-derived fungal strains for their potential to produce oxidases with laccase activities suitable for biotechnological applications. BMC Biotechnology, 20, 27. https://doi.org/10.1186/s12896-020-00617-y
Benkhaya, S., M’rabet, S., & Harfi, A. E. (2020). A review on classifications, recent synthesis and applications of textile dyes. Inorganic Chemistry Communications, 115, 107891.
https://doi.org/10.1016/j.inoche.2020.107891
Bu, R., Yan, B., Sun, H., Zhou, M., Bai, H., Cai, X., Mo, X., Su, G., & Jiang, C. (2021). Copper tolerance mechanism of the novel marine multi-stress tolerant yeast Meyerozyma guilliermondii GXDK6 as revealed by integrated omics analysis. Frontiers in Microbiology, 12, 771878.
https://doi.org/10.3389/fmicb.2021.771878
Couto, S. R. (2007). Decolouration of industrial azo dyes by crude laccase from Trametes hirsuta. Journal of Hazardous Materials, 148(3), 768–770. https://doi.org/10.1016/j.jhazmat.2007.06.123
D’Souza-Ticlo, D., Sharma, D., & Raghukumar, C. (2009). A thermostable metal-tolerant laccase with bioremediation potential from a marine-derived fungus. Marine Biotechnology, 11, 725–737.
https://doi.org/10.1007/s10126-009-9187-0
Dhouib, A., Hamza, M., Zouari, H., Mechichi, T., H’midi, R., Labat, M., Martinez, M. J., & Sayadi, S. (2005) Autochthonous fungal strains with high ligninolytic activities from Tunisian biotopes. African Journal of Biotechnology, 4, 431–436.
Domínguez, A., Gómez, J., Lorenzo, M., & Sanromán, Á. (2007). Enhanced production of laccase activity by Trametes versicolor immobilized into alginate beads by the addition of different inducers. World Journal of Microbiology and Biotechnology, 23, 367–373. https://doi.org/10.1007/s11274-006-9233-2
Ellouze, M., & Sayadi, S. (2016). White-rot fungi and their enzymes as a biotechnological tool for xenobiotic bioremediation. Intech, 17, 103–120. https://doi.org/10.5772/64145
Feng, Y., Cui, J., Xu, B., Jiang, Y., Fu, C., & Tan, L. (2023). A potentially practicable halotolerant yeast Meyerozyma guilliermondii A4 for decolorizing and detoxifying azo dyes and its possible halotolerance mechanisms. Journal of Fungi, 9(8), 851. https://doi.org/10.3390/jof9080851
Fernández‐Remacha, D., González‐Riancho, C., Osua, M. L., Arce, A. G., Montánchez, I., García‐Lobo, J. M., Estrada-Tejedor, R., & Kaberdin, V. R. (2022). Analysis of laccase‐like enzymes secreted by fungi isolated from a cave in northern Spain. Microbiology Open, 11, e1279. https://doi.org/10.1002/mbo3.1279
Flórez-Restrepo, M. A., García-Jiménez, M., López-Lugo, D. F., Múnera-Porras, L. M., Pino-Rodríguez, N. J., & Peñuela-Mesa, G. A. (2018). Discoloration of reactive black 5 by individual and consortium of Rhodotorula mucilaginosa, Galactomyces pseudocandidum and Escherichia coli free and immobilized. Ingeniería e Investigación, 38(3), 8-14. https://doi.org/10.15446/ing.investig.v38n3.68534
Gascuel, O. (1997). BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution, 14, 685–695. https://doi.org/10.1093/oxfordjournals.molbev.a025808
Gerritse, J., Leslie, H. A., de Tender, C. A., Devriese, L. I., & Vethaak, A. D. (2020). Fragmentation of plastic objects in a laboratory seawater microcosm. Scientific Reports, 10, 10945.
https://doi.org/10.1038/s41598-020-67927-1
Gharieb, M. M., Soliman, A. M., & Hassan, E. M. (2020). Biodegradation of the azo dyes Direct Red 81 and reactive brilliant red X-3B by wild strains of yeasts Meyerozyma guilliermondii and Naganishia diffluens. International Journal of Current Microbiology and Applied Sciences, 9(3), 1243-1260.
https://doi.org/10.20546/ijcmas.2020.903.145
Grossart, H.-P., Van den Wyngaert, S., Kagami, M., Wurzbacher, C., Cunliffe, M., & Rojas-Jimenez, K. (2019). Fungi in aquatic ecosystems. Nature Reviews Microbiology, 17, 339–354. https://doi.org/10.1038/s41579-019-0175-8
Hao, J., Song, F., Huang, F., Yang, C., Zhang, Z., Zheng, Y., & Tian, X. (2007). Production of laccase by a newly isolated deuteromycete fungus Pestalotiopsis sp. and its decolorization of azo dye. Journal of Industrial Microbiology & Biotechnology, 34, 233–240. https://doi.org/10.1007/s10295-006-0191-3
Hendricks, K. E., Christman, M. C., & Roberts, P. D. (2017). A statistical evaluation of methods of in-vitro growth assessment for Phyllosticta citricarpa: Average colony diameter vs. area. PLoS ONE, 12(1), e0170755. https://doi.org/10.1371/journal.pone.0170755
Jamee, R., & Siddique, R. (2019). Biodegradation of synthetic dyes of textile effluent by microorganisms: An environmentally and economically sustainable approach. European Journal of Microbiology and Immunology, 9, 114-118. https://doi.org/10.1556/1886.2019.00018
Jones, E. B. G., Pang, K.-L., Abdel-Wahab, M. A., Scholz, B., Hyde, K. D., Boekhout, T., Ebel, R., Rateb, M. E., Henderson, L., Sakayaroj, J., Suetrong, S., Dayarathne, M. C., Kumar, V., Raghukumar, S., Sridhar, K. R., Bahkali, A. H. A., Gleason, F. H., & Norphanphoun, C. (2019). An online resource for marine fungi. Fungal Diversity, 96, 347–433. https://doi.org/10.1007/s13225-019-00426-5
Juárez-Hernández, J., Castillo-Hernández, D., Pérez-Parada, C., Nava-Galicia, S., Cuervo-Parra, J. A., Surian-Cruz, E., Díaz-Godínez, G., Sánchez, C., & Bibbins-Martínez, M. (2021). Isolation of fungi from a textile industry effluent and the screening of their potential to degrade industrial dyes. Journal of Fungi, 7(10), 805.https://doi.org/10.3390/jof7100805
Kantharaj, P., Boobalan, B., Sooriamuthu, S., & Mani, R. (2017). Lignocellulose degrading enzymes from fungi and their industrial applications. International Journal of Current Research and Review, 9(21). https://doi.org/10.7324/ijcrr.2017.9211
Kaushik, P., & Malik, A. (2009). Fungal dye decolourization: Recent advances and future potential. Environment International, 35(1), 127–141. https://doi.org/10.1016/j.envint.2008.05.010
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549.
https://doi.org/10.1093/molbev/msy096
Kumar, V., Kaur, K., Gupta, G. K., & Sharma, A. K. (2013). Pyrazole containing natural products: Synthetic preview and biological significance. European Journal of Medicinal Chemistry, 69, 735-753.
https://doi.org/10.1016/j.ejmech.2013.08.053
Kyzas, G. Z., Fu, J., & Matis, K. A (2013). The change from past to future for adsorbent materials in treatment of dyeing wastewaters. Materials, 6, 5131–5158. https://doi.org/10.3390/ma6115131
Li, H., Zhang, W., Zhang, X., Tang, S., Men, P., Xiong, M., Li, Z., Zhang, Y., Huang, X., & Lu, X. (2022). Identification of PKS-NRPS hybrid metabolites in marine-derived Penicillium oxalicum. Marine Drugs, 20, 523. https://doi.org/10.3390/md20080523
Nakade, K., Nakagawa, Y., Yano, A., Konno, N., Sato, T., & Sakamoto, Y. (2013). Effective induction of pblac1 laccase by copper ion in Polyporus brumalis ibrc05015. Fungal Biology, 117, 52–61.
https://doi.org/10.1016/j.funbio.2012.11.005
Olufemi, A. G., Utieyin, O. O., & Adebayo, O. M. (2010). Assessment of groundwater quality and saline intrusions in coastal aquifers of Lagos metropolis, Nigeria. Journal of Water Resource and Protection,2,849-853.
Ozojiofor, U. O., Abdulsalami, M. S., Egbe, N. E., & Haroun, A. A. (2023). Fungi laccases: Structure, functions, and potential application in the biodegradation of pharmaceutical micropollutants. International Journal of Advanced Biotechnology and Research, 14(1), 24-38. http://doi.org/10.5281/zenodo.7697775
Palmieri, G., Giardina, P., Bianco, C., Fontanella, B., & Sannia, G. (2000). Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Applied and Environmental Microbiology, 66, 920–924. http://doi.org/10.1128/aem.66.3.920-924.2000
Piscitelli, A., Giardina, P., Lettera, V., Pezzella, C., Sannia, G., & Faraco, V. (2011). Induction and transcriptional regulation of laccases in fungi. Current Genomics, 12, 104–112.
http://doi.org/10.2174/138920211795564331
Raghukumar, C. (2008). Marine fungal biotechnology: An ecological perspective. Fungal Diversity,31,19–35.
Rao, M. A., Scelza, R., Scotti, R., & Gianfreda, L. (2010). Role of enzymes in the remediation of polluted environments. Journal of Soil Science and Plant Nutrition, 10(3), 333-353. http://doi.org/10.4067/S0718-95162010000100008
Rateb, M. E., & Ebel, R. (2011). Secondary metabolites of fungi from marine habitats. Natural Product Reports, 28, 290–344. http://doi.org/10.1039/c0np00061b
Reiss, R., Ihssen, J., Richter, M., Eichhorn, E., Schilling, B., & Thöny-Meyer, L. (2013). Laccase versus laccase-like multicopper oxidase: A comparative study of similar enzymes with diverse substrate spectra. PLoS One, 8(6), e65633. https://doi.org/10.1371/journal.pone.0065633
Richards, T. A., Jones, M. D. M., Leonard, G., & Bass, D. (2011). Marine fungi: Their ecology and molecular diversity. Annual Review of Marine Science, 4, 495–522. http://doi.org/10.1146/annurev-marine-120710-100802
Rosales, E., Pazos, M., & Sanroman, M. A. (2011). Comparative efficiencies of the decolourization of leather dyes by enzymatic and electrochemical treatments. Desalination, 278, 312–317.
https://doi.org/10.1016/j.desal.2011.05.041
Saravanakumar, K., & Kathiresan, K. (2014). Bioremoval of the synthetic dye malachite green by marine Trichoderma sp. SpringerPlus, 3, 631. https://doi.org/10.1186/2193-1801-3-631
Sharma, J., Sharma, S., & Soni, V. (2021). Classification and impact of synthetic textile dyes on Aquatic Flora: A review. Regional Studies in Marine Science, 45, 101802. https://doi.org/10.1016/j.rsma.2021.101802
Shin, K.-S., & Lee, Y-J. (2000). Purification and characterization of a new member of the laccase family from the white-rot basidiomycete Coriolus hirsutus. Archives of Biochemistry and Biophysics, 384, 109–115. https://doi.org/10.1006/abbi.2000.2083
Stielow, J. B., Lévesque, C. A., Seifert, K. A., Meyer, W., Iriny, L., Smits, D., Renfurm, R., Verkley, G. J., Groenewald, M., Chaduli, D., Lomascolo, A., Welti, S., Lesage-Meessen, L., Favel, A., Al-Hatmi, A. M., Damm, U., Yilmaz, N., Houbraken, J., Lombard, L., … Robert, V. (2015). One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia - Molecular Phylogeny and Evolution of Fungi, 35, 242–263. https://doi.org/10.3767/003158515X689135
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680. https://doi.org/10.1093/nar/22.22.4673
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A., Innis, D. H., Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR Protocols: A guide to methods and applications (pp. 315-322). Academic Press.
Yan, K., Zhang, Y., & Chi, Z. (2010). Distribution and diversity of Candida tropicalis strains in different marine environments. Journal of Ocean University of China, 9, 139–144.
Yang, P., Shi, W., Wang, H., & Liu, H. (2016). Screening of freshwater fungi for decolorizing multiple synthetic dyes. Brazilian Journal of Microbiology, 47, 828–834. https://doi.org/10.1016/j.bjm.2016.06.010
Yaseen, D. A., & Scholz, M. (2019). Textile dye wastewater characteristics and constituents of synthetic effluents: A critical review. International Journal of Environmental Science and Technology, 16,1193-1226.https://doi.org/10.1007/s13762-018-2130-z
Zhang, C., & Kim, S.-K. (2010). Research and application of marine microbial enzymes: Status and prospects. Marine Drugs, 8, 1920–1934. https://doi.org/10.3390/md8061920
Zouari-Mechichi, H., Mechichi, T., Dhouib, A., Sayadi, S., Martínez, A. T., & Martínez, M. J. (2006). Laccase purification and characterization from Trametes trogii isolated in Tunisia: Decolorization of textile dyes by the purified enzyme. Enzyme and Microbial Technology, 39, 141–148.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Suan Sunandha Rajabhat University

This work is licensed under a Creative Commons Attribution 4.0 International License.











